Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company. We are excited to be in business with you — email us at [email protected]
The ability to evolve new functions in living organisms is constrained by their fundamental mutation rate, which is the rate at which changes occur in the DNA sequence. This paper explores a novel method to accelerate the evolutionary process in E. coli, a common model organism used in various research fields. The authors developed a synthetic orthogonal replication system in E. coli, a system that operates independently of the host cell’s natural replication machinery. This system allows for targeted and accelerated evolution of specific DNA sequences without disrupting the genome.
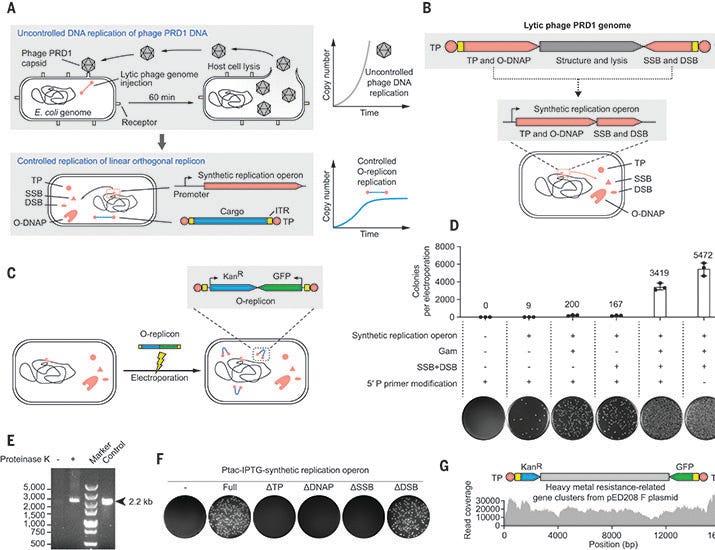
The cornerstone of this system is the orthogonal replicon, a linear piece of DNA containing a gene of interest and flanked by specific sequences that act as origins of replication. This replicon is not copied by the host cell’s DNA polymerases but is selectively replicated by an orthogonal DNA polymerase (O-DNAP), which is engineered to function independently of the host’s natural polymerases. The key innovation lies in the development of mutant O-DNAPs that can selectively increase the mutation rate of the orthogonal replicon, enabling rapid exploration of genetic variations.
To demonstrate the effectiveness of their system, the authors first established the stable inheritance of the orthogonal replicon in E. coli. They showed that the replicon could be maintained for hundreds of generations, a crucial requirement for any directed evolution strategy. The system was further optimized for its efficiency, allowing the integration of larger cargos (up to 16.5 kb) and enabling the coexistence of multiple orthogonal replicons within the same cell, paving the way for exploring complex genetic pathways.
The control of the orthogonal replicon copy number was achieved through the use of inducible promoters. The authors demonstrated the ability to adjust the copy number of the orthogonal replicon, and therefore the expression of genes encoded on it, over a wide dynamic range. This fine-tuning capability is crucial for controlling the evolutionary trajectory of specific genetic elements.
To assess the mutation rate of the orthogonal replicon, the authors performed fluctuation analysis, a technique that quantifies the frequency of mutations leading to a specific phenotype. They compared the mutation rates of the orthogonal replicon using wild-type O-DNAP and its engineered variants, demonstrating that the engineered O-DNAPs significantly increased the mutation rate of the orthogonal replicon without significantly altering the genome’s mutation rate. This selective increase in mutation rate is a key advantage, allowing targeted evolution without compromising the cell’s viability.
The authors then applied their system to accelerate the evolution of two functions: resistance to tigecycline, an antibiotic, and increased green fluorescence. By introducing an orthogonal replicon containing a tetracycline resistance gene and subjecting the cells to increasing concentrations of tigecycline, they successfully evolved resistance to 150 times higher concentrations of tigecycline compared to the initial gene. This process took only 12 days, highlighting the potential of this system for rapid evolution of antibiotic resistance.
Similarly, by introducing an orthogonal replicon containing a GFP gene and subjecting the cells to a selection process based on fluorescence, the authors evolved a variant of GFP that exhibited a thousandfold increase in fluorescence in just five days. This demonstrates the ability to evolve novel functionalities beyond antibiotic resistance, opening up possibilities for engineering various cellular functions.
This paper presents a robust platform for accelerating continuous evolution in E. coli, utilizing a synthetic orthogonal replication system. The key innovation lies in the controlled and independent evolution of specific DNA sequences within the cell, allowing for the rapid exploration of genetic diversity. This approach holds significant promise for accelerating the discovery and evolution of novel functions in various biological systems.
Further research is needed to explore the full potential of this system. For instance, investigating the application of this system to evolve more complex functionalities, such as metabolic pathways, could lead to the development of novel biotechnological applications. Moreover, expanding this system to other model organisms could open up new possibilities for understanding and manipulating various biological processes. The ability to accelerate evolution through synthetic biology holds immense promise for various research fields, from medicine to biotechnology and beyond, offering new pathways for solving complex challenges in the face of global challenges.
